Laboratoire Léon Brillouin
UMR12 CEA-CNRS, Bât. 563 CEA Saclay
91191 Gif sur Yvette Cedex, France
+33-169085241 llb-sec@cea.fr
Laboratoire Léon Brillouin
UMR12 CEA-CNRS, Bât. 563 CEA Saclay
91191 Gif sur Yvette Cedex, France
+33-169085241 llb-sec@cea.fr


|
To survive in their environment, bacteria must adapt to very different growth conditions. One way of doing this is by use of a protein called Hfq which forms complexes with specific small ribonucleic acids (RNAs). In a joint effort a CEA/IRAMIS/LLB group, associated with research groups from SOLEIL (DISCO beamline), CEA/I2BC, Curie Institute (Orsay) and ICP (Madrid), was able to show that the bacterial Hfq protein forms amyloid structures, such as those found in neurodegenerative diseases, and that these protein structures affect the integrity of bacterial membranes. For this research, various biophysical and biochemical approaches were used, including Small Angle scattering (SAXS and SANS), molecular Microscopy (cryo-TEM and AFM) and spectroscopy (SRCD on the DISCO beamline, and fluorescence). |
Bacteria are unicellular organisms, which constantly adapt to environmental changes. This adaptation is particularly critical when bacteria colonize a host, notably pathogenic bacteria, which adapt their metabolism depending on their growth conditions encountered within a host. Therefore they are able to quickly change their genetic expression system while communicating with each other. Although the way in which they adapt to their environment is not yet fully understood, it has recently been possible to show that bacteria use for this purpose a protein called Hfq and specific small RNAs which do not code for the expression of proteins, but do contribute to the regulation of gene expression [1]. Those two partners allow rapid changes of the bacterial genetic expression machinery, leading to metabolic adaptation of bacteria, e.g. in response to a drop in iron concentration levels.
A recent study, carried out by CEA/IRAMIS/LLB group, associated with research groups from SOLEIL (DISCO beamline), CEA/I2BC, Curie Institute (Orsay) and ICP (Madrid), shows that both Hfq protein and RNAs have the property of self-assembly in nanostructures [2,3]. The scientists notably showed that Hfq forms amyloid type assemblies similar to those found in neurodegenerative pathologies [4]. In bacteria nevertheless, these assemblies have a physiological function in contrast to eukaryotic amyloids. For the bacteria they are non-pathogenic, hence they are called functional amyloids.
Various biophysical and biochemical approaches have been used by the multidisciplinary team, including Small Angle X-ray or Neutron Scattering (SAXS/SANS), cryo Transmission Electron Microscopy (cryo-TEM) and Atomic Force Microscopy (AFM), and fluorescence or Synchrotron Radiation Circular Dichroism (SRCD) spectroscopy.
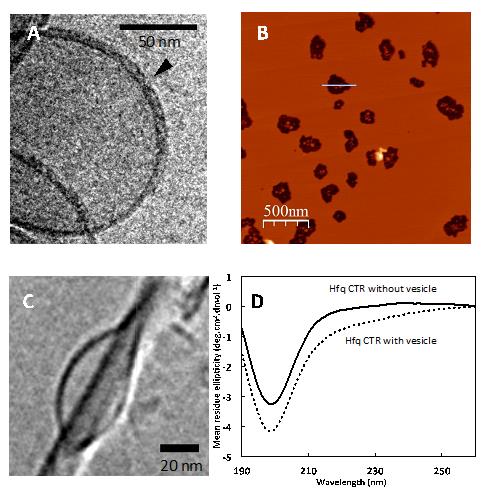
In addition the team has also identified that the assembled Hfq integrates in the bacterial membrane. Published in Scientific reports [5], these findings open novel perspectives on the way bacteria communicate with each other. It is well known that bacteria use rather small molecules to communicate with each other (in a process known as ‘quorum sensing') [6]. Based on these findings it seems likely that Hfq promotes the export of entire small nucleic acid molecules through the bacterial membrane by forming pores. This would allow direct genetic material exchange into the extracellular environment. This plausible hypothesis, based on the evidence of non-coding RNA found in the extracellular space, still needs to be proven by clarifying the molecular mechanism involved [7]. Ultimately, understanding this type of regulation could lead to a novel antibiotic development.
* Cryo-TEM : Imaging process by transmission electron microscopy after freezing, preserving the morphology and structure of biological molecules. The Nobel Prize in Chemistry 2017 was awarded to the scientists at the origin of this technique.
Références :
[1] Hfq and its constellation of RNA
J. Vogel & B.F. Luisi, Nat Rev Microbiol 9, 578-589 (2011)
[2] Multiple approaches for the investigation of bacterial small regulatory RNAs self-assembly
C. Lavelle, F. Busi, & V. Arluison, Methods Mol Biol 1297, 21-42 (2015).
[3] Techniques to analyse sRNA protein cofactor self-assembly in vitro"
D. Partouche, A. Malabirade, T. Bizien, M. Velez, S. Trepout, S. Marco, V. Militello, C. Sandt, F. Wien and V. Arluison, Methods Mol Biol (2017) in press.
[4] New insight into the structure and function of Hfq C-terminus
E. Fortas, F. Piccirilli, A. Malabirade, V. Militello, S. Trépout, S. Marco, A. Taghbalout A, V. Arluison Biosci Rep 35 (2015).
[5] Membrane association of the bacterial riboregulator Hfq and functional perspectives
A. Malabirade, J. Morgado-Brajones, S. Trépout, F. Wien, I. Marquez, J. Seguin, S. Marco, M. Velez & V. Arluison. Sci Rep 7, 10724 (2017).
[6] Comprehensive analysis reveals how single nucleotides contribute to noncoding RNA function in bacterial quorum sensing
S.T. Rutherford, J.S. Valastyan, T. Taillefumier, N.S. Wingreen & B.L. Bassler
Proc Natl Acad Sci U S A 112, E6038-6047.
[7] The extracellular RNA complement of Escherichia coli
Ghosal A, B.B. Upadhyaya, J.V. Fritz, A. Heintz-Buschart, M.S. Desai, D. Yusuf, D. Huang, A. Baumuratov, K. Wang , D. Galas, P. Wilmes, Microbiologyopen (2015).
See the highlight from the Synchrotron SOLEIL: Functional amyloids and bacterial adaptation.
Contact CEA-IRAMIS : Véronique Arluison (LLB)
Collaborations :
•  Institut Rayonnement Matière de Saclay • Laboratoire Léon Brillouin (LLB)
Institut Rayonnement Matière de Saclay • Laboratoire Léon Brillouin (LLB)
• Biologie et Systèmes Désordonnés - GBSD (appareils 7C2, Paxe, Mibemol et Muse)
